4-D Genomic Dynamics In Ecology Evolution
&

“Education is not the filling of a pail, but the lighting of a fire.” ~WILLIAM BUTLER YEATS



4-D Genomic Dynamics In Ecology & Evolution
Our group focuses on the genetic and regulatory basis of adaptation and population dynamics in natural species and cell lines by integrating ecological and evolutionary theory, population genetics, system biology approaches and multi-omics methods. For example, we are asking how the life-history tradeoffs shape the evolutionary processes and the formation of genetic diversity in different species, and what the consequences are in terms of adaptation or resistance in metapopulations. We are also interested in the interact and co-evolution of genomic variations and three-dimensional (3D) genome organization and their effects on adaptation.
Featured Publication

May 5, 2025
Evolutionary divergence on the Qinghai-Tibet Plateau: How life-history traits shape the diversity of plateau zokor and pika populations.
研究表明,即便在相似的生态背景下,不同物种由于内在特征的差异,仍可能呈现出截然不同的群体进化轨迹与局域适应策略,提示研究者在探讨遗传结构与群体多样性的形成机制时,应关注物种内在特性与外部环境之间的交互作用。该发现为传统群体遗传学议题,尤其是关于物种形成过程的Wright-Fisher模型相关争论,提供了新的视角与启示。
Apr 7, 2023
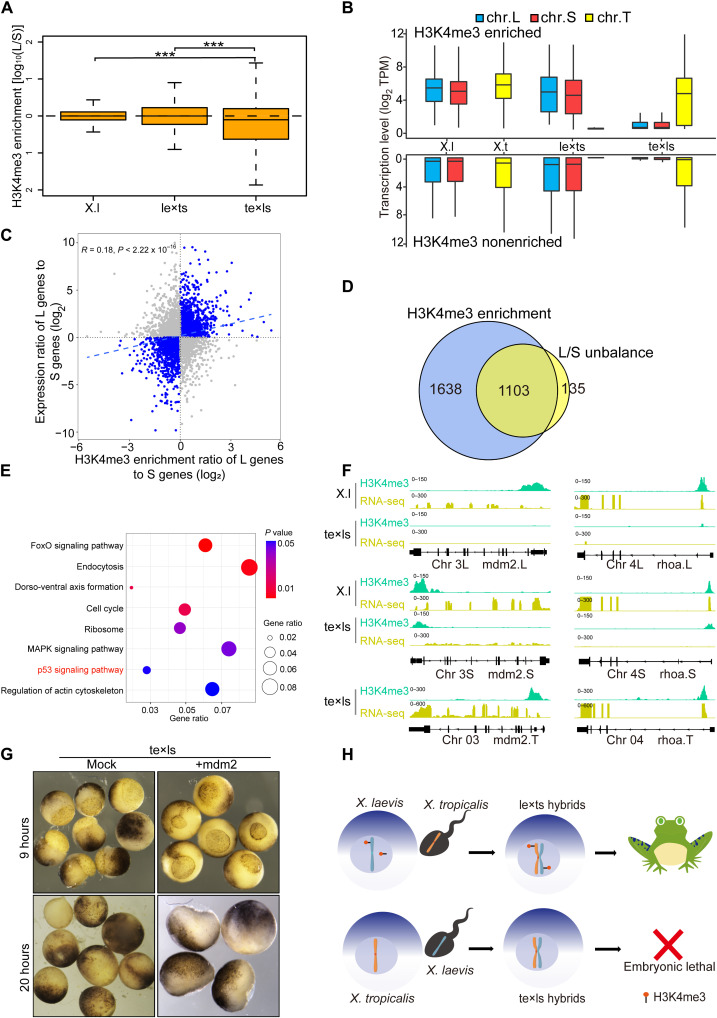
Modification of Maternally Defined H3K4me3 Regulates the Inviability of Interspecific Xenopus Hybrids
本研究利用两种爪蟾的独特的种间杂交体系,进一步探讨了种间杂交中染色质不亲和性的机制。该研究强调了母系定义的H3K4me3修饰,通过P53信号通路调控胚胎早期发育,在生殖隔离和维持杂交种亚基因组平衡的重要性。研究非洲爪蟾杂交种并破译物种形成的机制将加深我们对生殖隔离、物种进化和调节早期胚胎发育表观遗传修饰的理解。
Jan 7, 2022
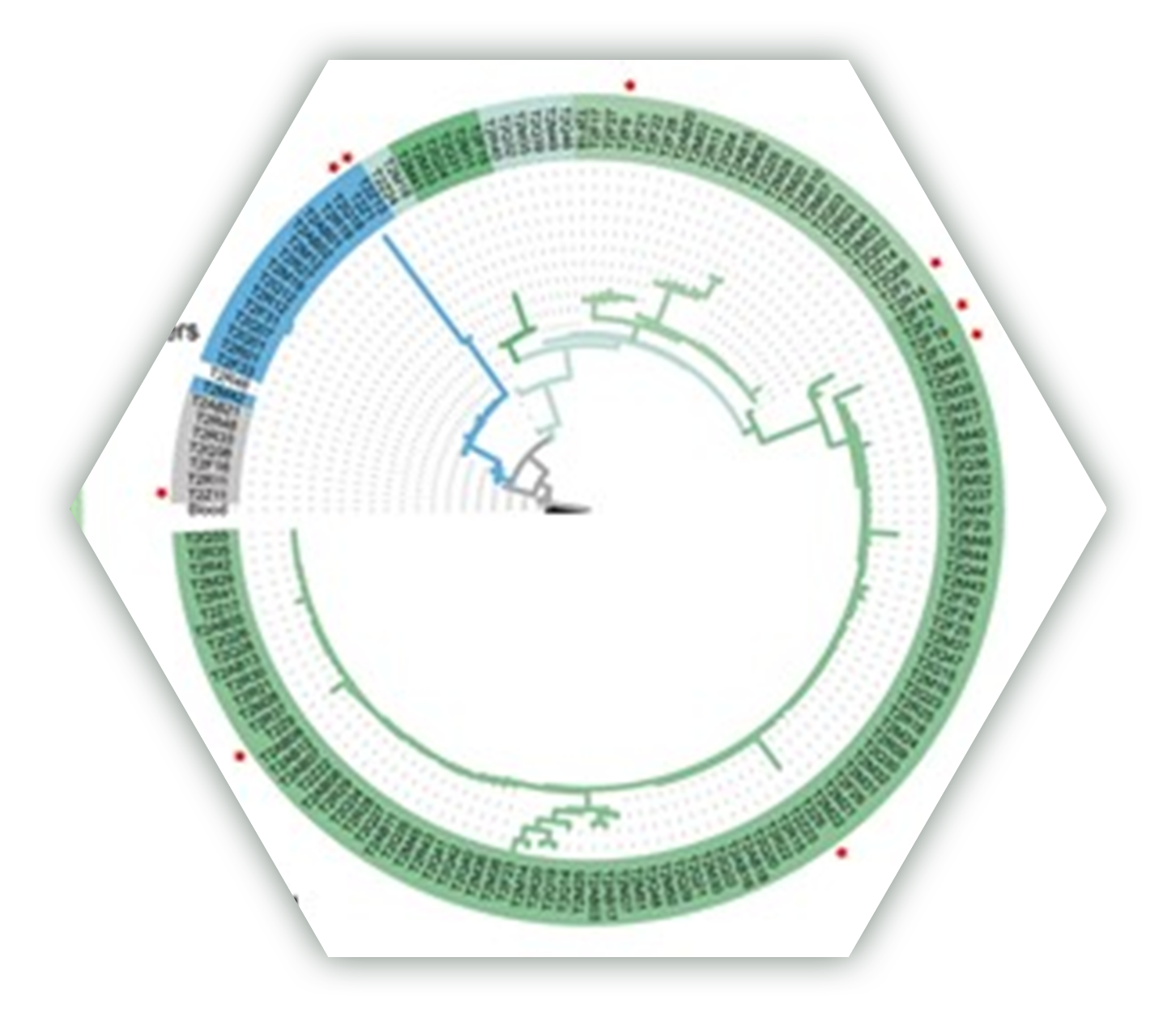
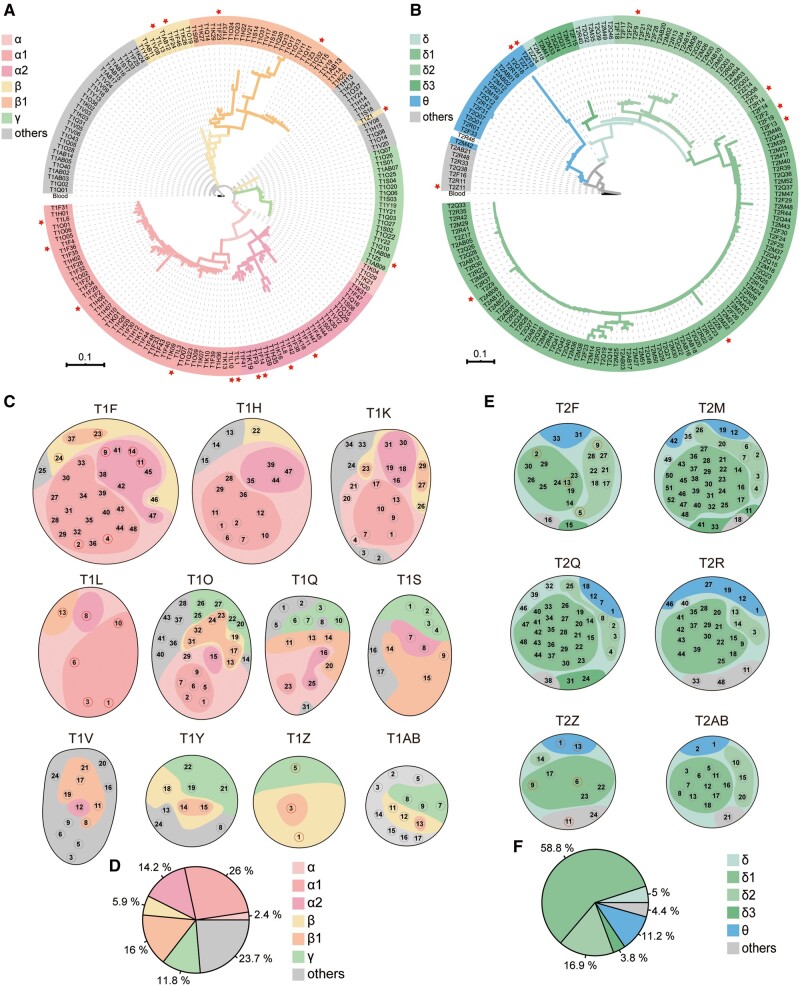
Evolution under Spatially Heterogeneous Selection in Solid Tumors
结构化种群的进化驱动力和动态进程及其预测:检测结构化群体进化过程中的自然选择颇具挑战性,不同阶段起主导作用的驱动力有所不同。我们引入中性空间模型,揭示了中性进化和自然选择的交替作用,驱动了肿瘤内遗传结构、克隆多样性的形成以及肿瘤的克隆扩张和取代。
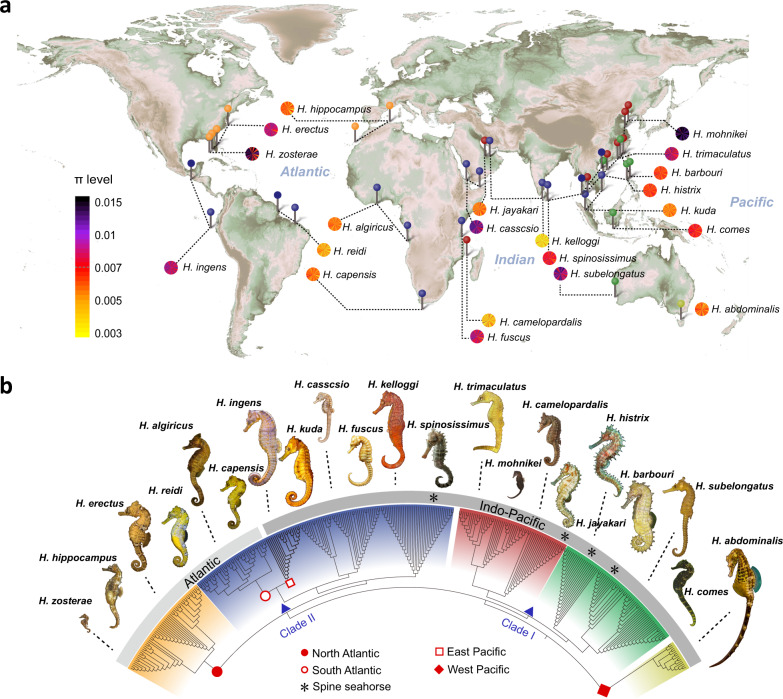
Feb 17, 2021
Genome sequences reveal global dispersal routes and suggest convergent genetic adaptations in seahorse evolution
该研究以全球优势海马类群为研究对象,首次阐明其起源中心及在全球层面的扩散模式与时空路径特征,并揭示了海马种化过程中的性状演化机制;为全球海洋生物的扩散及其多样性演化研究提供了重要的研究范式,为探索大洋生物资源从印-太海域向大西洋迁移与演化规律等研究提供了科学理论支撑。

Nov 24, 2015
Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution
为了检测瘤内进化是否符合达尔文进化或非达尔文进化,我们选取了一种肝癌细胞系,通过深度采样,利用全外显子测序和基因型检测,以中性模型为基础,建立肿瘤细胞群体遗传学模型并作理论模拟。我们发现高度的遗传多样性、突变克隆的生长和空间与突变-漂变中性模型相吻合,没有发现选择驱动内肿瘤多样性的证据,群体内部的亚克隆之间没有适应性差异,肿瘤治疗应关注非达尔文进化过程。





 云南省昆明市龙欣路17号
云南省昆明市龙欣路17号